
Thus, RPCs contain CMG along with Mcm10, Ctf4, Pol α, Mrc1, Csm3, Tof1, FACT, and Topo I. Indeed, epitope tagging of CMG subunits, followed by cell extract pull-outs and mass spectrometry, has identified a large ‘replisome progression complex’ (RPC) that contains CMG along with several other factors, some of which are essential for cell viability ( Gambus et al., 2006 Gambus et al., 2009). While in vitro synthesis of the leading strand replisome has been accomplished with the purified CMG complex from budding yeast ( Georgescu et al., 2014), the discontinuous lagging strand is a much more difficult process and the number of proteins required for lagging strand synthesis is currently unknown. In addition, many replication fork-associated proteins undergo modifications in response to the cell cycle or DNA damage. Numerous other proteins travel with eukaryotic replication forks and have no bacterial homolog or known function. The 11-subunit CMG complex consisting of the Mcm2-7 ‘motor’, a GINS heterotetramer, and one Cdc45 subunit ( Moyer et al., 2006 Ilves et al., 2010 Costa et al., 2011 Costa et al., 2014) provides the helicase activity. Priming is performed by Pol α, a 4-subunit enzyme that contains an RNA primase and DNA polymerase activity and makes short RNA-DNA hybrid primers of 25–35 nucleotides ( Kaguni and Lehman, 1988 Waga and Stillman, 1998 Benkovic et al., 2001 Stillman, 2008). Cellular studies reveal that eukaryotes use two different DNA polymerases for the leading and lagging strands, Pols ε and δ, respectively ( Lee et al., 1989 Weinberg and Kelly, 1989 Tsurimoto et al., 1990 Waga and Stillman, 1998 Benkovic et al., 2001 Pursell et al., 2007 Kunkel and Burgers, 2008 Nick McElhinny et al., 2008 Stillman, 2008). Important future studies must now address how the replisome deals with obstacles created by certain DNA-binding proteins and damaged DNA and how it interfaces with the molecules that control cell division and DNA repair.Ĭomposition of the eukaryotic replisome and the function of its various proteins is an area of active investigation. then found that the eukaryotic polymerases are actively prevented from copying the ‘wrong’ strand of DNA and suggest that the helicase enzyme that unwinds the DNA might be behind this activity. This exclusivity is unique to eukaryotic DNA replication, as bacterial polymerases can use either DNA strand as a template. Further experiments revealed that the polymerase enzyme that operates on the leading strand cannot work on the lagging strand and vice versa.
have essentially rebuilt a eukaryotic replisome from 31 different proteins in a test tube and confirmed that it can make both leading and lagging DNA strands-just like in a normal cell. The replisomes in bacterial cells have been well studied, but many researchers are investigating the composition of the replisome in animals, plants, and fungi (collectively called eukaryotes).
#DIFFERENCE BETWEEN OKAZAKI FRAGMENT AND LAGGING STRAND SERIES#
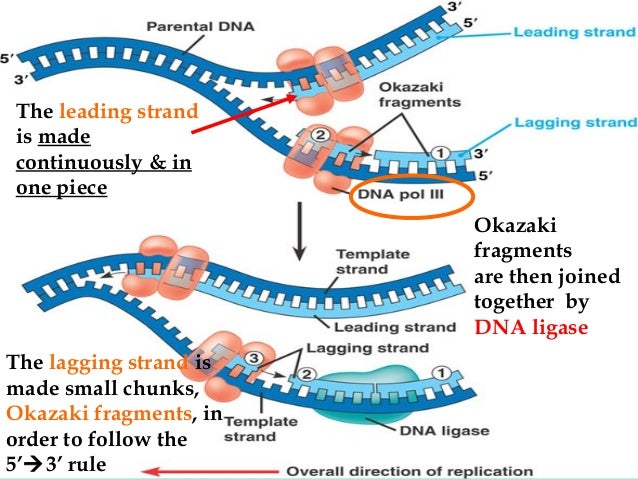
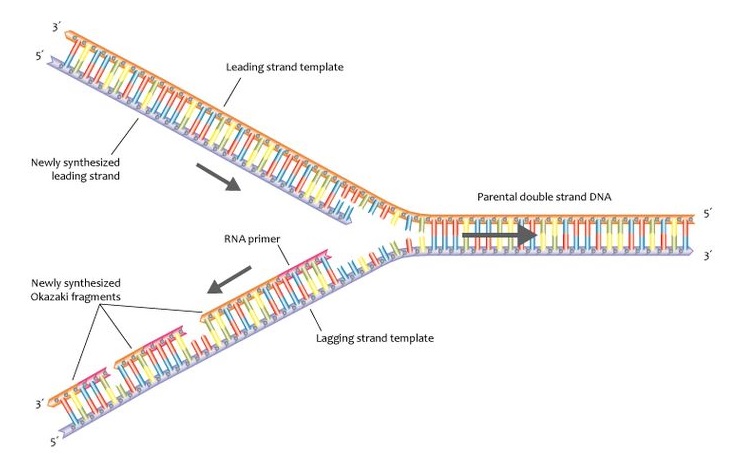
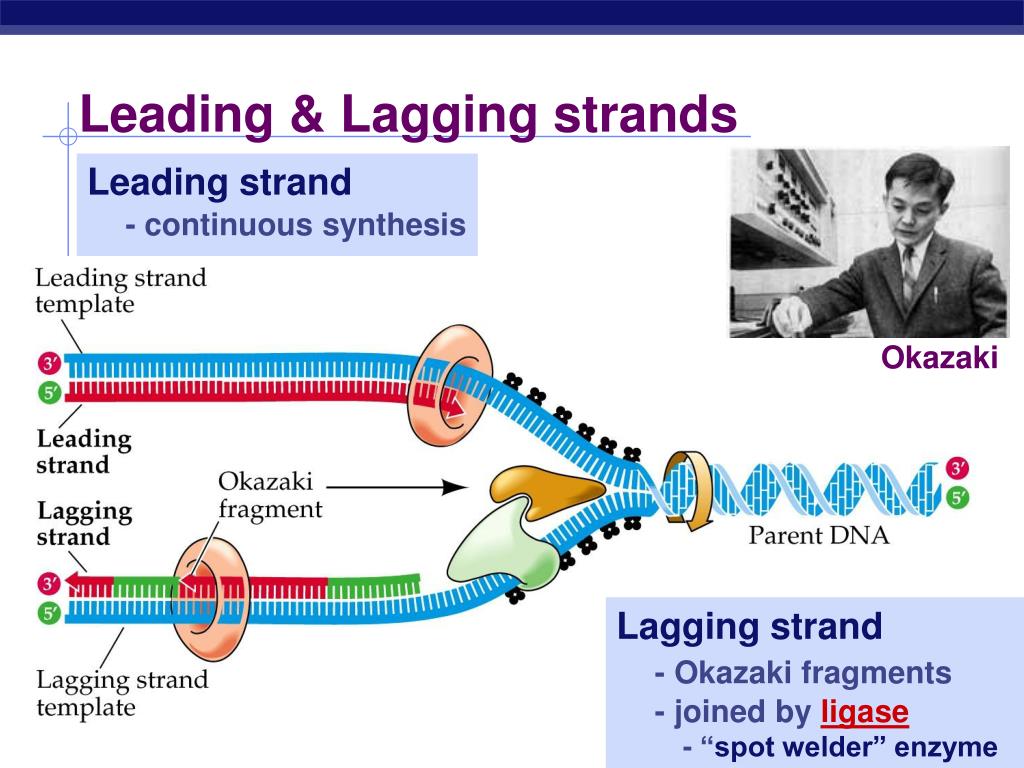
The other newly forming strand-the ‘lagging strand’-is made in the opposite direction, as a series of short fragments that are later joined together. One of these new strands is built continuously and called the ‘leading strand’. This process continues while other enzymes, called polymerases, use the exposed single strands as templates to make complementary new strands of DNA. First, an enzyme called a helicase starts to unwind the double-stranded DNA into two single strands. DNA replication requires complex molecular machinery called a replisome, which comprises multiple proteins, enzymes, and other molecules.
Cells must replicate their DNA before they divide so that the newly formed cells can each receive a copy of the same genetic material.


 0 kommentar(er)
0 kommentar(er)
